On novel inserts in SARS-nCoV-2
There’s been a lot of debate about the Pradhan paper1 : a paper that had a short life span on the bioRxiv pre-print server a while back – Jan. 31, 2020 – to be precise. It showed the presence of four unique inserts in the SARS-nCoV-2 genome that are not present in any other coronaviruses, including SARS-nCoV (I might refer to the original SARS as ‘original recipe chicken’ in this article, or some variation of this). “How did they get there?” was the question of the day. And “Why was the paper really withdrawn by the authors”? Will they rise to their intentions to ‘revise it in response to comments received from the research community on their technical approach and their interpretation of the results’? What does that even mean? And who exactly is this ‘research community’?
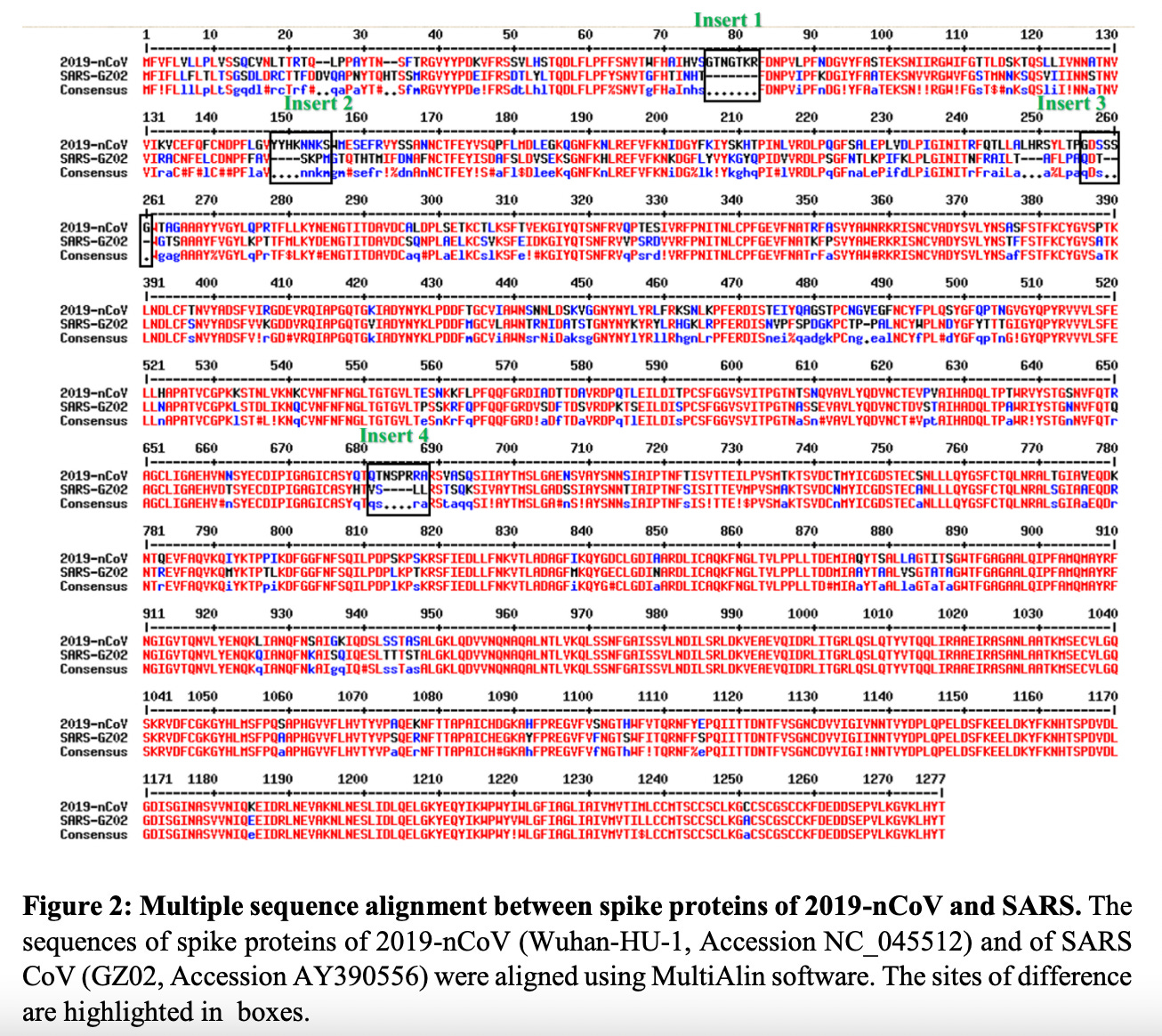
Figure 1: Sequence alignment between SARS and SARS-2 spike proteins showing the 4 inserts. https://doi.org/10.1101/2020.01.30.927871
I am part of the research community and I have independently confirmed the presence of these 4 inserts in SARS-2 that are not present in the original recipe chicken SARS.

Figure 2: Alignment of NC_045512.2 SARS-nCoV-2 with AY390556 SARS original chicken using MEGA software.
All four of these inserts had sequence similarity to motifs found in the Human Immunodeficiency Virus (HIV). It was my impression that most people bought the idea that there was no possible way that function could be ascribed to these tiny little peptides. But then again, the fact that they were all so beautifully situated on the spike protein for easy access as binding sites (potentially functional), does indeed raise some furry eyebrows. The manuscript was very rapidly withdrawn from pre-print by the authors themselves. Someone did not want this paper being read. So, please read it. It’s not long and it’s not complicated.
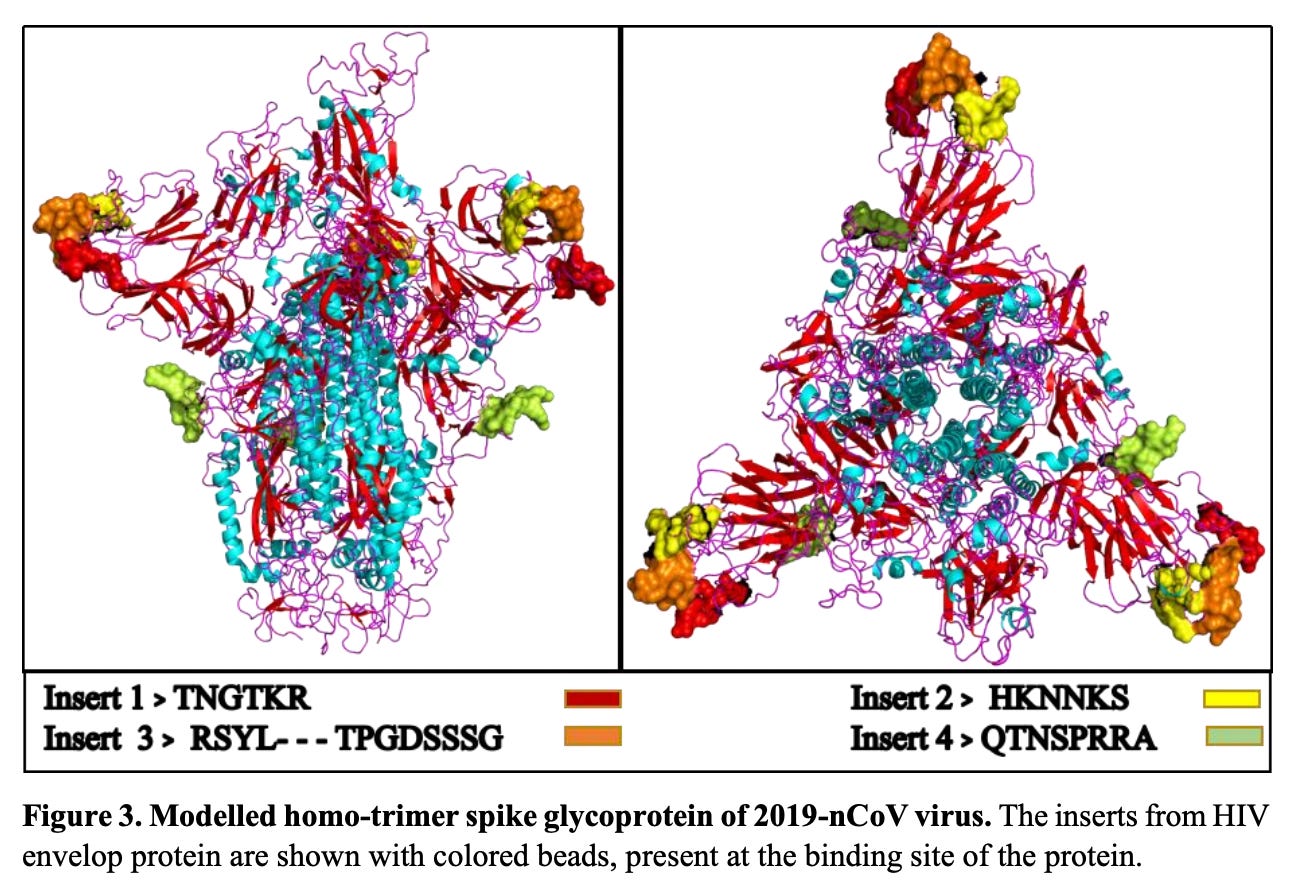
Figure 3: Exposed peptides – great spots with respect to binding-ability. https://doi.org/10.1101/2020.01.30.927871
It was clear to me as soon as I saw the sequence alignment of SARS-nCoV-2 and SARS-nCoV (Figure 1) that something was ‘artificially’ different about this new version of SARS. My exact words (that I uttered in tears as I read the Pradhan paper) were: “They made it”. I had hoped to be wrong and I let it play on my mind while keeping an eye on supportive publications hitting the pre-print servers or heaven forbid, published with or without retraction.2
Una pausa: How easily did we get to a place where retractions are common in specific areas of science? Retractions are a big deal, and they typically come with shame due to issues of fraud, scientific misconduct or conflicts of interest. The large number of retractions in the COVID era also flies in the face of the fact that the rate of retractions over the past few decades has been declining.3 It would be interesting to compare and contrast the number of retractions to the number of published articles on COVID-19 and SARS-2 currently listed on Pubmed. According to a Pubmed search using keywords ‘COVID-19’ or ‘SARS CoV-2’, there are 478,363 articles currently published on Pubmed. This is just the tip of the iceberg, but it is safe to say, that there are hundreds of thousands of peer-reviewed articles on this subject matter published to date. The rate of retraction as of 2018, according to the above footnote (2) was 4 in 10,000. I leave this to one of my brilliant colleagues to hash out.
At this point in my investigations, I am pretty convinced that these 4 inserts were put there with intelligent intension. Let me tell you, I would love to get my hands on the lab notebooks of the people who did these experiments. With great admiration and an equal amount of disrespect, I woefully shame these people for not ensuring that their work did not fall into the wrong hands, and/or for not ensuring that their brilliant work was not used for nefarious purposes. I know that we can’t control certain applications of our research entirely, but I personally know very high profile people who have been approached by certain ‘high-ranking’ entities to buy their research only to be told: ‘yeah, that would be a NO’. It can be done. Just saying.
On nuclear translocation of spike proteins and spike mRNA: P to the RRARSV
Please refer to a recently published pre-print entitled: “Nuclear translocation of spike mRNA and protein is a novel pathogenic feature of SARS2 CoV-2”.4 This paper is not yet peer-reviewed but the results are very interesting and support the construction hypothesis, in my opinion. It also validates and, in fact, vindicates yet another seminal retracted paper of the COVID era published in 2021 in the journal Viruses entitled: “SARS-CoV-2 spike impairs DNA damage repair and inhibits V(D)J recombination in vitro”.5 I have written about this paper here and Arkmedic wrote this entire story up comprehensively and has archived this work as well here.
The main point of interest in this latest paper which has yet to be withdrawn from the pre-print server (we’re waiting), is that the sequence of amino acids thought to be acting solely as a furin-cleavage site in the SARS-nCoV-2 spike protein, has actually been shown to be a component part in a motif that assists in the translocation of the spike protein and SARS-CoV-2 S mRNA, to the nucleus of infected cells. This transpires via a Nuclear Location Signal (NLS) motif on the spike protein that is unique among human pathogenic beta-coronaviruses and a novel pathogenic feature of SARS-CoV-2. The question in my mind becomes, does the full-length spike modified mRNA template encode this NLS? I recently read an absolutely excellent Substack article written by Joomi on the subject matter of spike products – as in, what the hell are these modified mRNA products actually being translated into once injected into people? – that you can read here. I also wrote about this ages ago and you can read that here.
It is disturbing to me that a transmembrane type 1 glycoprotein (you know, the sticky-outy bit on the outside of the viral sphere) whose function is to enable viral entry into cells, is shipped to the nuclei of infected cells. What are the implications of this? What exactly is the result of this with regard to pathogenicity? Does the redistribution of the spike from the membrane surface to the nuclear membrane surface affect immune recognition? We don’t know yet. We do know that these shots are messing up a lot of people physiologically. Is this primarily due to the proteins that actually end up being translated? Does the NLS motif always get translated? Does variability in NLS translation-ability explain the range of adverse events seen clinically? What are the implications of injecting the modified coding material of the spike protein, repeatedly, into live subjects?
The authors also show that the NLS is functional as a nuclear transporter in infected cells in the context of spike protein-spike mRNA complexes. This means that with the help of spike, the mRNA is also getting trafficked to the nucleus. What are the implications for systemic intracellular distribution of spike and spike mRNA? What are the implications with regard to integration into the human genome? This remains to be seen.
I fully believe that the integration paper is being written right now as you read this.
As we found similar intracellular distribution of both S mRNA and S protein, we hypothesized that S protein interacts with S mRNA to translocate the protein‒mRNA complex to different subcellular locations, including the cytoplasm and nucleus, but not the cell surface.
Ultimately, the authors don’t make any definitive conclusions about the construction hypothesis, but the fact that this NLS exists in this version of SARS-2 and not in the original recipe SARS, is, when combined with the other spike peptides that shouldn’t be there, suspicious, to say the least.
While our result does provide direct evidence for the presence of the NLS motif and nuclear translocation of the S protein, our results do not confirm nor deny that the NSPR sequence has a natural origin. Instead, our results showed that the inserted sequence NSPR was a functional NLS motif, which increased the intracellular distribution of the S protein, including novel nuclear translocation. The novel nuclear translocation of the SARS CoV-2 S protein suggests that: 1. the nuclear translocation of the S protein reduces its surface expression, but whether it contributes to evading host immune recognition remains to be determined; and 2. the colocalization of the S protein with S mRNA suggests that the S protein has an RNA binding motif, which remains to be determined.
So the NLS in SARS-nCoV-2 wouldn’t be there unless this PRRA insert had been introduced. That’s the bottom line. How did it get there? The will of God? Nature? Or perhaps idiotic humans playing God?
In conclusion, the SARS-CoV-2 S protein has a functional pat7 NLS “PRRARSV”, that results in one out of four S proteins translocating into the nucleus in infected cells. S Protein appears to shuttle S mRNA (possibly the genome) into the nucleus as well. Thus, the NLS of the S protein may contribute to the evasion of the host immune response and is a novel pathogenic feature of SARS-CoV-2.
Here is a beautiful confocal image (Figure 4 bottom from paper) showing nuclear colocalization of spike-protein-spike mRNA complexes as seen by the tiny little white dots. It’s hard to see, but it’s there.
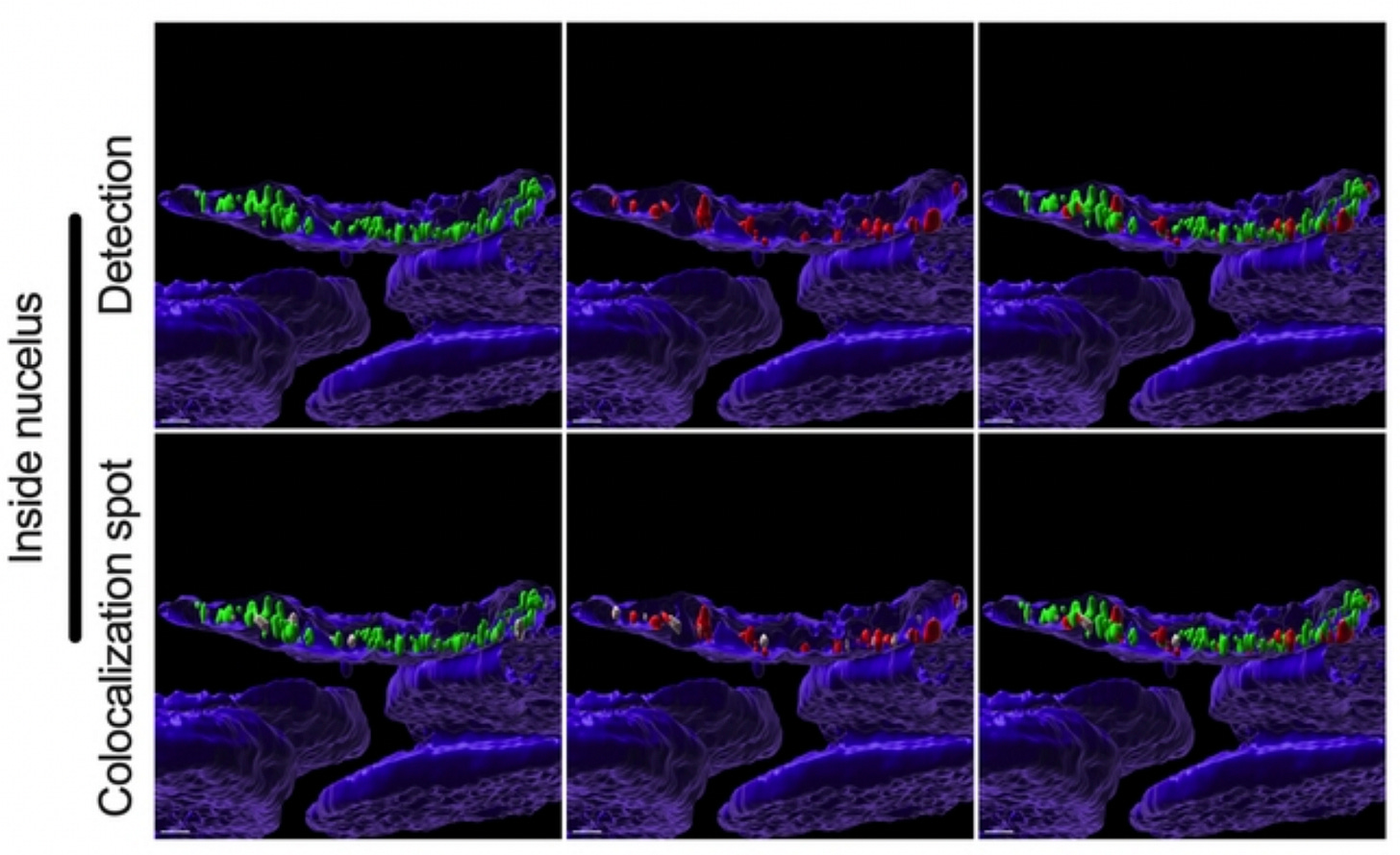
Figure 4: Fig 4. Colocalization between S mRNA and S protein inside infected cells. The images (see 457 Fig. 3) were analyzed by using the surface rendering and colocalization features of IMARIS. S protein and S mRNA distribution and colocalization in the cytoplasm (top panel), on the nuclear surface (middle panel) and inside the nucleus (bottom panel). The specific region of colocalization is indicated by a white spot. Scale bar 0.5 µm. https://www.biorxiv.org/content/10.1101/2022.09.27.509633v1
Since this article addresses the construction hypothesis of the SARS-2 spike protein, before I move onto another recent publication review, I would like to point to another great write-up by Scoops McGoo about the possibility that our infamous SARS-2 was made in Canada. Yes, you read that right. If the so-called ‘Winnipeg lab documents’ are eventually un-redacted and handed over to Parliament (and the public) via lawful processes and actions, it will quite likely tip the scales to induce a course-change on this entire COVID-19 narrative.
On flappy flaps of SARS-CoV-2
Another interesting paper that I want to include in this article is one entitled: “Flap structure within receptor binding domain of SARS-CoV-2 spike periodically obstructs hACE2 Binding subdomain bearing similarities to HIV-1 protease flap”6published in Scientific Reports Nature portfolio on September 22, 2022 by Michael H. Peters. You can read a summary Substack by John Paul here. Many thanks to the author for this fascinating work and our short email exchanges.
This paper is quite interesting in that the author makes a comparison between the HIV-1 protease flap and a sequence similar structure in the RBD of the SARS-nCoV-2 . He describes the latter as ‘a dynamic flap that opens and closes the dominant energetic binding subdomain of the SARS-2 spike protein’. This flippy-flappy bit found in HIV-1 and SARS-2 is meant to an intermediary of sorts between binding and not binding. If it’s open/unobstructed: binding can occur. That is, binding between the RBD of the SARS-nCoV-2 spike protein and human ACE-2. I see it as a kind of bouncer for binding. You know that big tattooed guy with his arms crossed at the doorway to the bar? That guy is the flap. And his arms are really flexible and move really fast. The RBD is the bar and we, the as-of-yet drunken punk rock show goers, are the ACE-2.
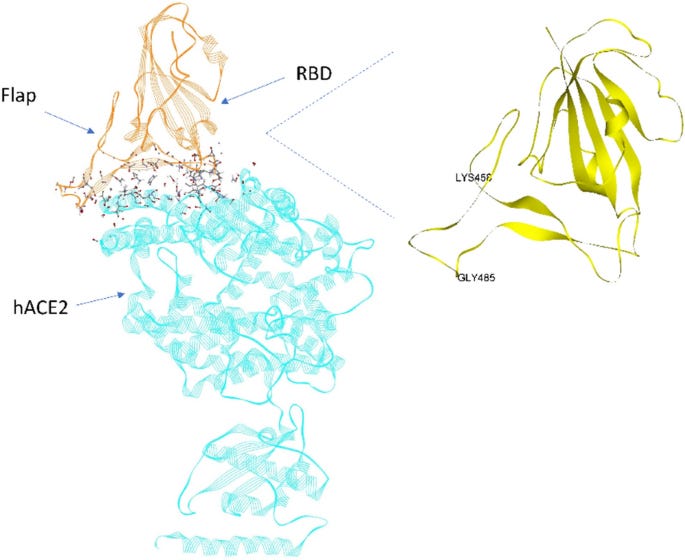
Figure 5: Figure 1: Dominant energetic atom–atom interactions (shown in sticks) between SARS-CoV-2 RBD and full length hACE2. A dynamic flap is also shown in this figure which opens and closes the dominant energetic binding subdomain of SARS-CoV-2 spike protein, as detailed herein. https://www.nature.com/articles/s41598-022-20656-z.
The RBD flap in the Up state protomer periodically obstructs the binding site on an approximate 70 ns time interval and is reminiscent of an HIV-1 protease polypeptide flap that opens and closes to modulate that enzymes activity.
More specifically, the SARS-2 flippy-flappy bit is part of a hair-pin loop789 that is involved in ACE-2 binding. This would be like if the bouncer was permanently holding… Nunchaku. The Nunchaku would be the hair-pin. I think.

The author proposes that [antibodies] or cyclic urea inhibitors10 could be used therapeutically to prevent binding of spike and ACE-2 by (blocking hair-pin opening and) keeping the flap closed. This is vital information with regard to prevention of infection and clearance of spike from the human body and is perhaps the most important take home message of this work.
The flap may have potential as a therapeutic target and bears dynamic and sequence similarity to the HIV-1 protease polypeptide flap, although its possible modulating role in SARS-CoV-2 binding is unknown and no claims are made here in that regard.
The RBD has a flap structure that is not involved in the primary binding hACE2 subdomain. The flap (loop) itself is part of a beta hairpin loop structure that contains turns and beta strands that are associated with the primary hACE2 binding residues of the RBD.
The author maintains that although the sequence similarity and time interval of obstruction/unobstruction dynamics between the flippy-flaps of HIV-1 and SARS-2 is clear, he cannot as yet draw conclusions as to the functionality or modulating role of this flap. But seeing as how he correctly provides evidence of the role of the HIV-1 protease peptide that acts in much the same way,1112 it makes perfect sense to me that the SARS flap on the RBD is playing a functional role with regard to binding.
It is also important to point out that this flappy-flap is also found on SARS original chicken recipe which you can see in blue in Figure 7 below. I gotta say, I LOVE this diagram I made using Pymol.
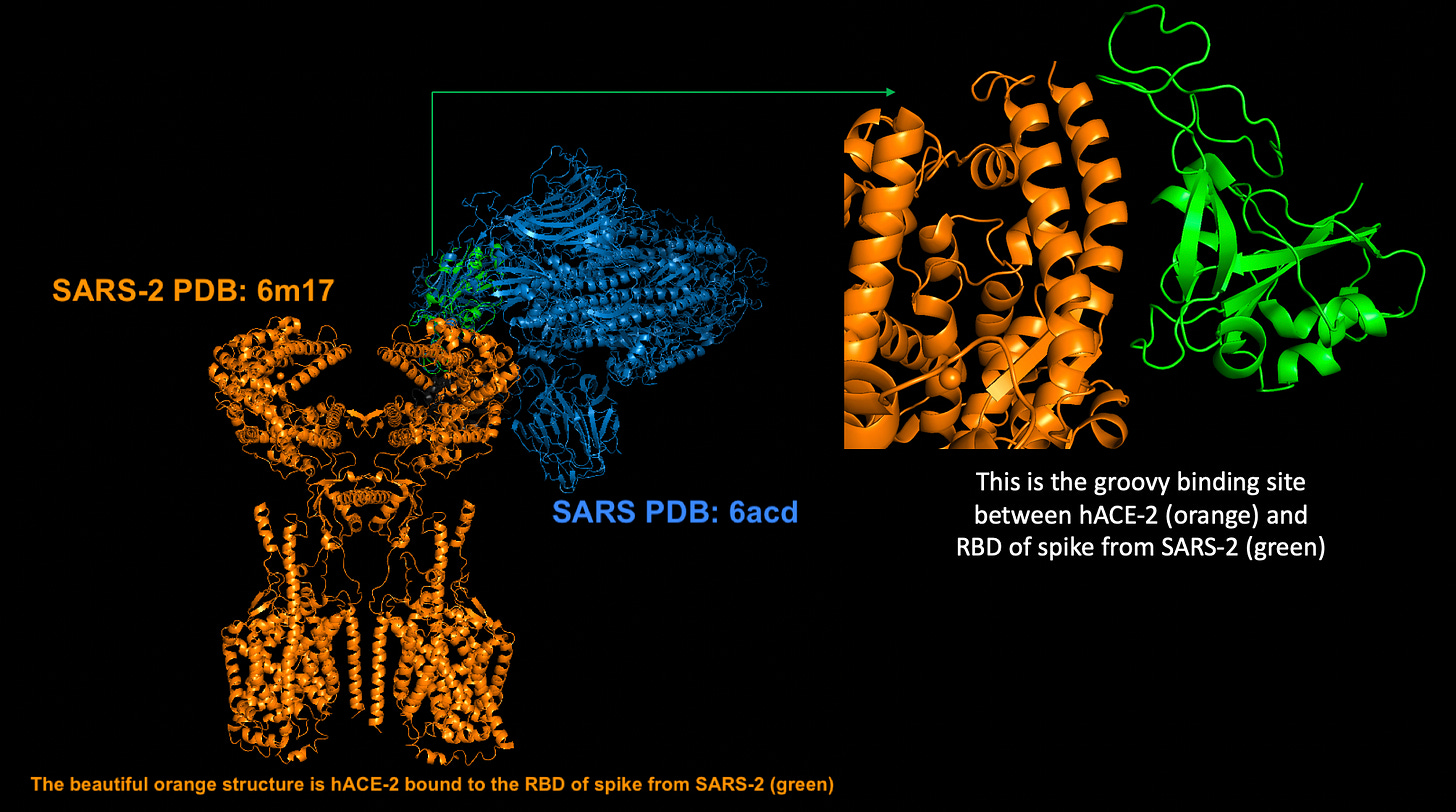
Figure 7: ACE-2:SARS-2 spike crystal structure in bound form (PDB: 6m17 – orange and green) aligned with SARS (PDB: 6acd – blue).
I am left with 2 burning questions:
- Was the SARS-nCoV-2 (spike) constructed by the hands of man?
- Why the consistent disappearance of both peer-reviewed and pre-print articles on this subject matter?
Is it not the very duty of us as scientists to fight come hell or high-water to ensure that many points of views are allowed to be examined as par for the scientific discovery course? Is it not our job to have open discourse and debate? Do we make mistakes? Sure. That’s why discussion is so vital! That’s why we have peer review.
The peer-review process is, in theory, a brilliant idea. To ensure that scientific works that get published are of the highest quality; this is essential! Somewhere along the line, perhaps even by design, something went wrong and it became a money-driven popularity contest by my observation, and I think everybody in the scientific has felt this way at one point or another. Why are scientists charged exorbitant fees to have their peers review their work? This never has made sense to me, and it seems that COVID has beautifully and ironically revealed the very dirty and broken system that is the current peer review process machine. Sad as this is, we have to know the problem before we can remedy it. If you want a short video talk by Kevin McKernan that describes a solution away from the current peer review system, go here.
We cannot continue to allow bureaucrats a seat at the science table. And we cannot allow them to insert themselves into doctor patient relationships – like some gross point mutation that induces mis-folding. That’s pretty much what has happened. Everything in our systems is mis-folding right now. I hope you guys enjoy reading this as much as I enjoyed writing it.
- Prashant Pradhan, Ashutosh Kumar Pandey, Akhilesh Mishra, Parul Gupta, Praveen Kumar Tripathi, Manoj Balakrishnan Menon, James Gomes, Perumal Vivekanandan, Bishwajit Kundu. Uncanny similarity of unique inserts in the 2019-nCoV spike protein to HIV-1 gp120 and Gag. bioRxiv 2020.01.30.927871; doi: https://doi.org/10.1101/2020.01.30.927871
- My opinions are my own and not reflect any others’ opinions including the authors of the articles herein.
- https://www.science.org/content/article/what-massive-database-retracted-papers-reveals-about-science-publishing-s-death-penalty
- Sarah Sattar, Juraj Kabat, Kailey Jerome, Friederike Feldmann, Kristina Bailey, Masfique Mehedi. Nuclear translocation of spike mRNA and protein is a novel pathogenic feature of SARS-CoV-2. bioRxiv preprint doi: https://doi.org/10.1101/2022.09.27.509633
- Jiang, H.; Mei, Y.-F. SARS–CoV–2 Spike Impairs DNA Damage Repair and Inhibits V(D)J Recombination In Vitro. Viruses 2021, 13, 2056. https://doi.org/10.3390/v13102056.
- Peters, M.H. Flap structure within receptor binding domain of SARS-CoV-2 spike periodically obstructs hACE2 Binding subdomain bearing similarities to HIV-1 protease flap. Sci Rep 12, 16236 (2022). https://doi.org/10.1038/s41598-022-20656-z
- https://en.wikipedia.org/wiki/Beta_hairpin
- Dyer RB, Maness SJ, Peterson ES, Franzen S, Fesinmeyer RM, Andersen NH. The mechanism of beta-hairpin formation. Biochemistry. 2004 Sep 14;43(36):11560-6. doi: 10.1021/bi049177m. PMID: 15350142.
- Chan KW, Luo CC, Lu H, Wu X, Kong XP. A site of vulnerability at V3 crown defined by HIV-1 bNAb M4008_N1. Nat Commun. 2021 Nov 9;12(1):6464. doi: 10.1038/s41467-021-26846-z. PMID: 34753944; PMCID: PMC8578649.
- Hodge CN, Aldrich PE, Bacheler LT, Chang CH, Eyermann CJ, Garber S, Grubb M, Jackson DA, Jadhav PK, Korant B, Lam PY, Maurin MB, Meek JL, Otto MJ, Rayner MM, Reid C, Sharpe TR, Shum L, Winslow DL, Erickson-Viitanen S. Improved cyclic urea inhibitors of the HIV-1 protease: synthesis, potency, resistance profile, human pharmacokinetics and X-ray crystal structure of DMP 450. Chem Biol. 1996 Apr;3(4):301-14. doi: 10.1016/s1074-5521(96)90110-6. PMID: 8807858.
- https://pubs.rsc.org/en/content/articlehtml/2003/ob/b208248a
- https://www.sciencedirect.com/topics/medicine-and-dentistry/hiv-1-protease
Source – https://jessicar.substack.com/p/it-turns-out-that-the-prrarsv-motif
